Ish Venkatesh
Research Assistant Professor
Marquette University
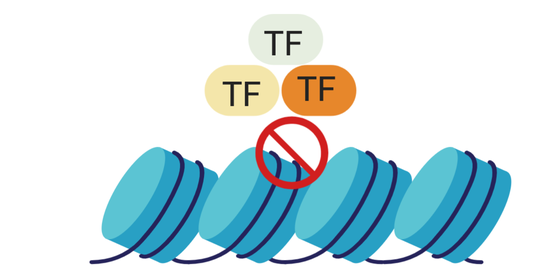
My research revolves around understanding molecular mechanisms that control axon growth, with the ultimate goal of using this information to induce axon regeneration following injury. I have spent the last decade tackling the problem of why injured neurons in the central nervous system fail to upregulate the right sets of genes supportive of axon re-growth, which ultimately determines regenerative success. I have identified and addressed transcriptional and epigenetic barriers to re-growth using a combination of methods including Bioinformatics, Functional Genomics, and High-Content Screening. I am currently looking to transition to a tenure track position in India in the coming year.
Interests
- Axon regeneration
- Spinal Cord Injury
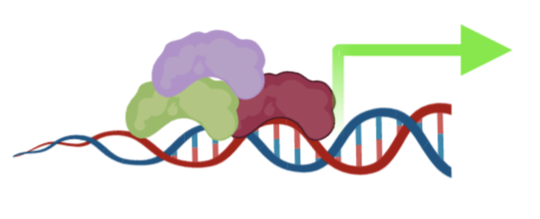
- Gene regulation
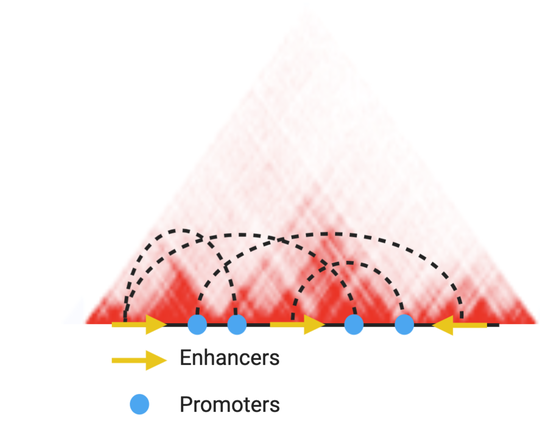
- 3D Genomics
Education
PhD in Neuroscience, 2014
University of Wisconsin, Milwaukee
Bachelors in Biotechnology, 2009
Anna University